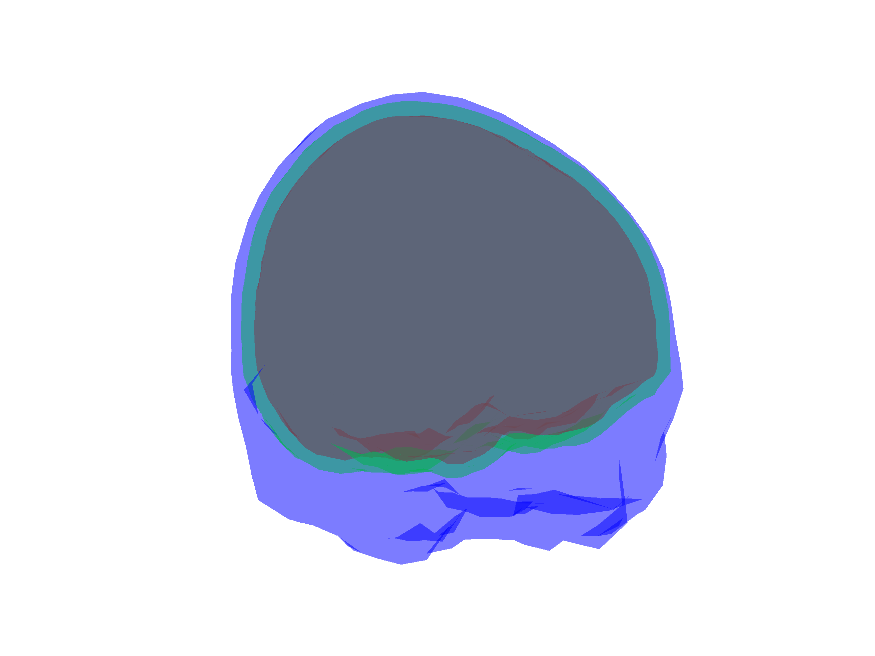
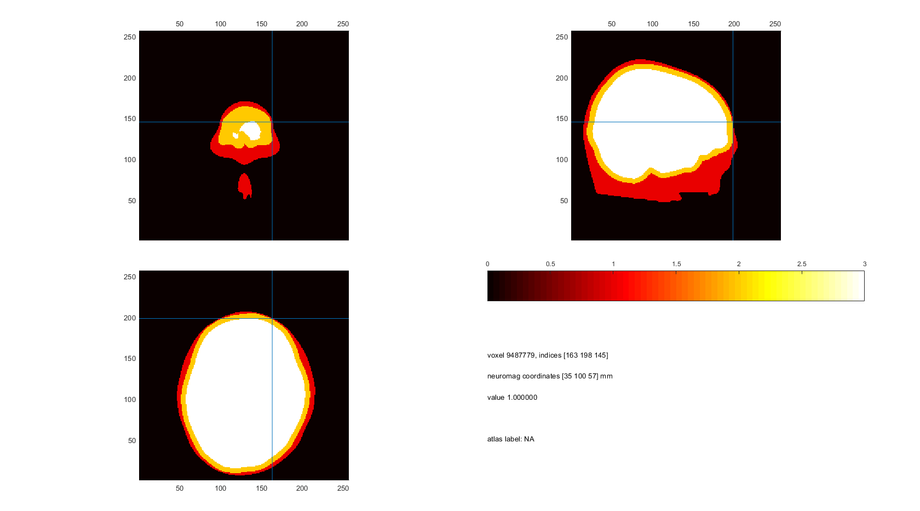
Output of ft_volumesegment:
the input is volume data with dimensions [256 256 256]
using 'OldNorm' normalisation
Smoothing by 0 & 8mm..
Coarse Affine Registration..
Fine Affine Registration..
performing the segmentation on the specified volume, using the old-style segmentation
SPM12: spm_preproc (v4916) 19:07:24 - 17/12/2020
========================================================================
Completed : 19:12:22 - 17/12/2020
creating scalpmask ... using the anatomy field for segmentation
smoothing anatomy with a 5-voxel FWHM kernel
thresholding anatomy at a relative threshold of 0.100creating brainmask ... using the summation of gray, white and csf tpms
smoothing brainmask with a 5-voxel FWHM kernel
thresholding brainmask at a relative threshold of 0.500
creating skullmask ... using the brainmask
the call to "ft_volumesegment" took 394 seconds
Output of ft_prepare_headmodel:
reordering the boundaries to: 3 2 1
your surface normals are inwards oriented
your surface normals are inwards oriented
your surface normals are inwards oriented
om_assemble version 2.4.1 compiled at Aug 22 2018 19:47:28 using OpenMP
Executing using 8 threads.
| ------ om_assemble
| -HM
| C:\Users\Jason\AppData\Local\Temp\ft_om_171220191925304\om.geom
| C:\Users\Jason\AppData\Local\Temp\ft_om_171220191925304\om.cond
| C:\Users\Jason\AppData\Local\Temp\ft_om_171220191925304\hm.bin
| -----------------------
(DEPRECATED) Please consider updating your geometry file to the new format 1.1 (see data/README.rst): C:\Users\Jason\AppData\Local\Temp\ft_om_171220191925304\om.geom
This geometry is a NESTED geometry.
Info:: Mesh name/ID : 1
# vertices : 800
# triangles : 1596
Euler characteristic : 2
Min Area : 5e-06
Max Area : 0.000156791
Info:: Mesh name/ID : 2
# vertices : 1600
# triangles : 3196
Euler characteristic : 2
Min Area : 7.5e-06
Max Area : 5.75348e-05
Info:: Mesh name/ID : 3
# vertices : 2400
# triangles : 4796
Euler characteristic : 2
Min Area : 6.40312e-06
Max Area : 4.01746e-05
Info:: Domain name : air
Conductivity : 0
Composed by interfaces : +1
Considered as the outermost domain.
Interface "1"= { mesh "1"(outermost) }
Info:: Domain name : scalp
Conductivity : 0.333333
Composed by interfaces : +2 -1
Interface "2"= { mesh "2" }
Interface "1"= { mesh "1"(outermost) }
Info:: Domain name : skull
Conductivity : 0.004167
Composed by interfaces : +3 -2
Interface "3"= { mesh "3" }
Interface "2"= { mesh "2" }
Info:: Domain name : brain
Conductivity : 0.333333
Composed by interfaces : -3
Interface "3"= { mesh "3" }
!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!
!!!!!!!!!!! WARNING !!!!!!!!!!!
!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!
2 meshes are intersecting !
Info:: Mesh name/ID : 2
# vertices : 1600
# triangles : 3196
Euler characteristic : 2
Min Area : 7.5e-06
Max Area : 5.75348e-05
Info:: Mesh name/ID : 3
# vertices : 2400
# triangles : 4796
Euler characteristic : 2
Min Area : 6.40312e-06
Max Area : 4.01746e-05
Error using ft_headmodel_openmeeg (line 186)
Aborting OpenMEEG pipeline due to above error.
Error in ft_prepare_headmodel (line 313) headmodel = ft_headmodel_openmeeg(geometry, 'conductivity', cfg.conductivity, 'isolatedsource',
cfg.isolatedsource, 'tissue', cfg.tissue);
Code:
%% Segment the brain
cfg = [];
cfg.output = {'brain' 'skull' 'scalp'};
mri_segmented = ft_volumesegment(cfg, mri_resliced);
%% Creating meshes
cfg = [];
cfg.method = 'projectmesh';
cfg.tissue = 'brain';
cfg.numvertices = 2400;
mesh_brain = ft_prepare_mesh(cfg, mri_segmented);
mesh_brain = ft_convert_units(mesh_brain, 'm'); % Use SI Units
cfg = [];
cfg.method = 'projectmesh';
cfg.tissue = 'skull';
cfg.numvertices = 1600;
mesh_skull = ft_prepare_mesh(cfg, mri_segmented);
mesh_skull = ft_convert_units(mesh_skull, 'm'); % Use SI Units
cfg = [];
cfg.method = 'projectmesh';
cfg.tissue = 'scalp';
cfg.numvertices = 800;
mesh_scalp = ft_prepare_mesh(cfg, mri_segmented);
mesh_scalp = ft_convert_units(mesh_scalp, 'm'); % Use SI Units
mesh_eeg = [mesh_brain mesh_skull mesh_scalp];
%% Plot mesh
figure
ft_plot_mesh(mesh_brain, 'edgecolor', 'none', 'facecolor', 'r')
ft_plot_mesh(mesh_skull, 'edgecolor', 'none', 'facecolor', 'g')
ft_plot_mesh(mesh_scalp, 'edgecolor', 'none', 'facecolor', 'b')
alpha 0.3
view(132, 14)
print('-dpng', fullfile(fdir,'meshes.png'))
%% Troubleshoot meshes
mri_segmented_binary = mri_segmented; % make a copy
binary_brain = mri_segmented.brain;
binary_skull = mri_segmented.skull | mri_segmented.brain;
binary_scalp = mri_segmented.scalp | mri_segmented.skull | mri_segmented.brain;
mri_segmented_binary.combined = binary_scalp + binary_skull + binary_brain;
cfg = [];
cfg.funparameter = 'combined';
cfg.location = [-76 -27 32];
ft_sourceplot(cfg, mri_segmented_binary);
%% Head models (component 1)
cfg = [];
cfg.method = 'openmeeg';
cfg.conductivity = [1 1/80 1] *(1/3); % S/m
headmodel = ft_prepare_headmodel(cfg, mesh_eeg);
headmodel = ft_convert_units(headmodel, 'm'); % Use SI Units